Wastewater sampling has become a popular way over the years to keep track of the health of a population, including human ones, as pathogens are often detectable in the effluence from toilets. Since most houses connected to the centralized sewer systems, this means that a few sampling sites suffice to keep tabs on which viruses are circulating in an area. While sampling this wastewater is easy, the actual RNA analysis using PCR (polymerase chain reaction) still has to be performed in laboratories, adding complex logistics. An approach for on-site analysis using microfluidics was tested out by [Yuwei Pan] et al., as recently published in Cell.
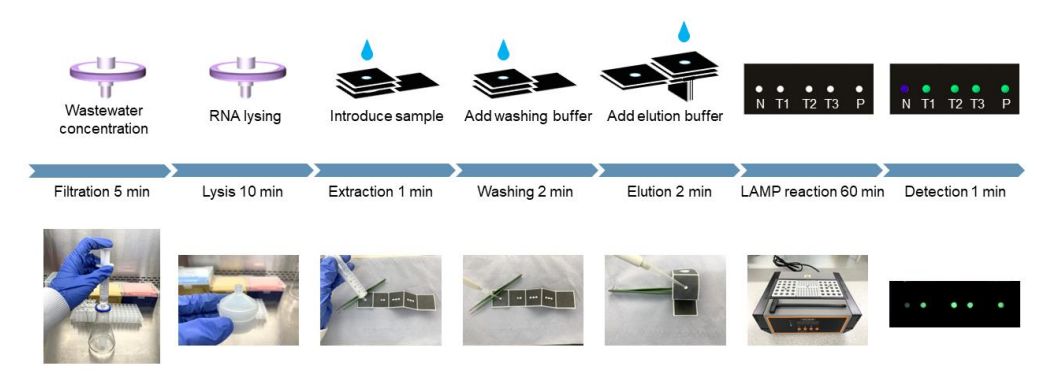
This particular approach uses RT-LAMP (reverse-transcription loop-mediated isothermal amplification) to increase the amount of genetic material, which has the significant benefit over PCR that it does not require multiple thermal cycles, instead being run at a constant temperature. The filter paper used as the basis has wax microchannels printed on it, which help to guide the filtered wastewater to the reaction chambers. This is in many ways reminiscent of the all too familiar linear flow self-tests (RAT: rapid antigen test) that have become one of the hallmarks of the SARS-CoV-2 pandemic.
What this paper microfluidic device adds is that it doesn’t merely contain antigens, but performs the lysis (i.e. breakdown of the virus particles), genetic material multiplication using RT-LAMP and subsequent presence detection of certain RNA sequences to ascertain the presence of specific viruses. Having been used in the field already since 2020 in the UK, the researchers envision this type of on-site analysis to be combined with a smartphone for instant recording and transmission to health authorities.
Some of the benefits of this approach would be lower cost, easier logistics and faster results compared to shipping wastewater samples to central laboratories.